Editing promoter regions using CRISPR-Cas9 technology can regulate gene expression to different levels at the transcriptional level and produce a large number of unpredictable quantitative trait variations. But it would take a lot of effort to screen out the ideal mutant. Therefore, the development of new methods to accurately and predictably regulate gene expression could greatly expand the existing gene expression regulation toolbox and provide strong technical support for crop genetic improvement.
Upstream open reading frame (uORF) is a common regulatory element of translation in eukaryotic mrnas, and it can inhibit the translation of primary open reading frame (pORF).
In 2018, Caixia Gao's research team at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences took the lead in editing uORF using CRISPR-Cas9 technology, established a method to finely up-regulate endogenous gene translation, and used this method to cultivate lettuce germplasm with significantly increased vitamin C content. In 2020, Gao Caixia's research team applied this technology to the genetic improvement of strawberry, and obtained a series of new strawberry germplasm with graded sugar.
On March 9, 2023, Gao Caixia's team published a paper entitled: Tuning plant phenotypes by precise, graded downregulation of gene expression. Based on previous studies, we developed a novel method to accurately down-regulate the expression of target gene proteins by designing upstream open reading frame (uORF), which provides an important technical means for future molecular design breeding.

The length of upstream open reading frame (uORF) and the distance between uORF and pORF will affect the inhibition of uORF on pORF translation. Therefore, the research team hypothesized that the translation of target genes could be inhibited by the following two strategies: one is to generate a new uORF from the beginning in the 5' untranslated region (5'UTR) of the target gene; The second is to mutate the stop codon of the endogenous uORF in situ to extend its expression frame length. The results of protoplast transient system showed that these two strategies could effectively inhibit pORF translation to different levels, and had little effect on mRNA expression. Later, the research team used base editing and guided editing systems to obtain rice mutant plants containing newly created UorFs or extended endogenous UorFs. By detecting the protein expression level and phenotype of the mutant plants, it was found that the effects of the introduced uORF variant on the expression and phenotype of the target protein were consistent with the results of the transient system.
In order to achieve continuous gradient downregulation of target gene expression, this study combined the above two strategies to design a series of uORFs with different inhibitory abilities in the 5’utr region of rice OsTCP19, OsTB1 and OsDLT genes, respectively. Transient system results showed that the translation level of pORF was reduced to 2.5% to 84.9% of the original level in a gradient way, which realized the gradient knockdown of gene. OsDLT genes encode members of the GRAS protein family, which are involved in rice brassinolide signal transduction pathway and regulate many important agronomic traits such as plant height, tiller number and seed size. In this study, a group of mutants with different leaf Angle, plant height and tiller number were obtained by editing the 5’utr of OsDLT gene as the target, and the phenotypic change trend of the mutants was consistent with the instantaneous system prediction.
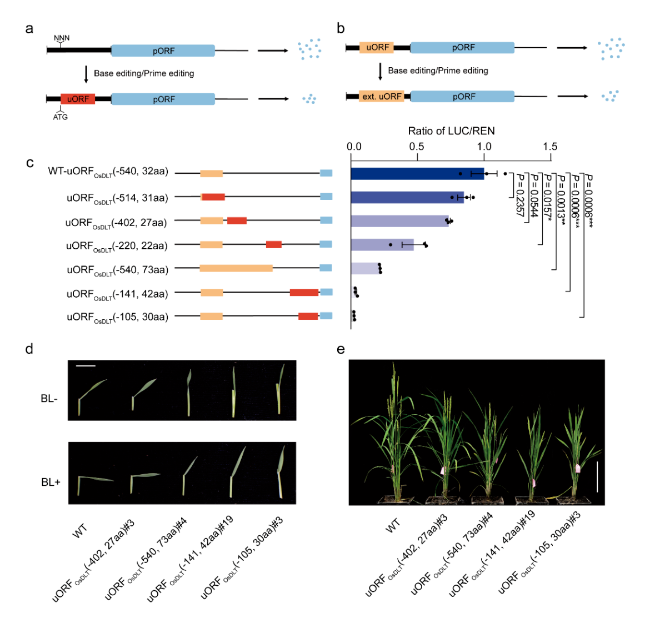
Rice plants with target traits were cultivated by editing uORF. (a, b) Create new uORFs (a) from scratch or extend endogenous uORFs (b) to inhibit protein expression of target genes. (c) down-regulation of uORFs by producing a series of gradients with different inhibitory capacities. (d, e) Susceptibility to brassinosteroids (d) and plant phenotype (e) of a mutant containing the uORFs variant.
In conclusion, this study has developed a universal and predictable method to fine-regulate gene expression through the design of uORF, which provides an important technical means for future molecular design and breeding.
Chen Tao Xue, a postdoctoral fellow in Gao Caixia's research group, is the first author of the paper, and Caixia Gao is the corresponding author. This research was supported by the National Key Research and Development Program, the Strategic Key Project of the Chinese Academy of Sciences and the Ministry of Agriculture and Rural Affairs of China.
Article from: biological exploration, if there is infringement, please contact delete.


 Contact Us
Contact Us
 Return
Return